Have you ever wondered how bacteria can generate nano-motors that can reach speeds of over 100,000 revolutions per minute? Or how our cells build tiny generators that power our body through all our daily tasks?
Through billions of years of evolution and natural selection, nature has been fabricating such diverse and complex structures effortlessly with high precision, adaptability and robustness [1]. This occurs through a biological phenomenon known as ‘self-assembly’ where many brainless molecules somehow self-organise and spontaneously form functional machinery with emergent properties.
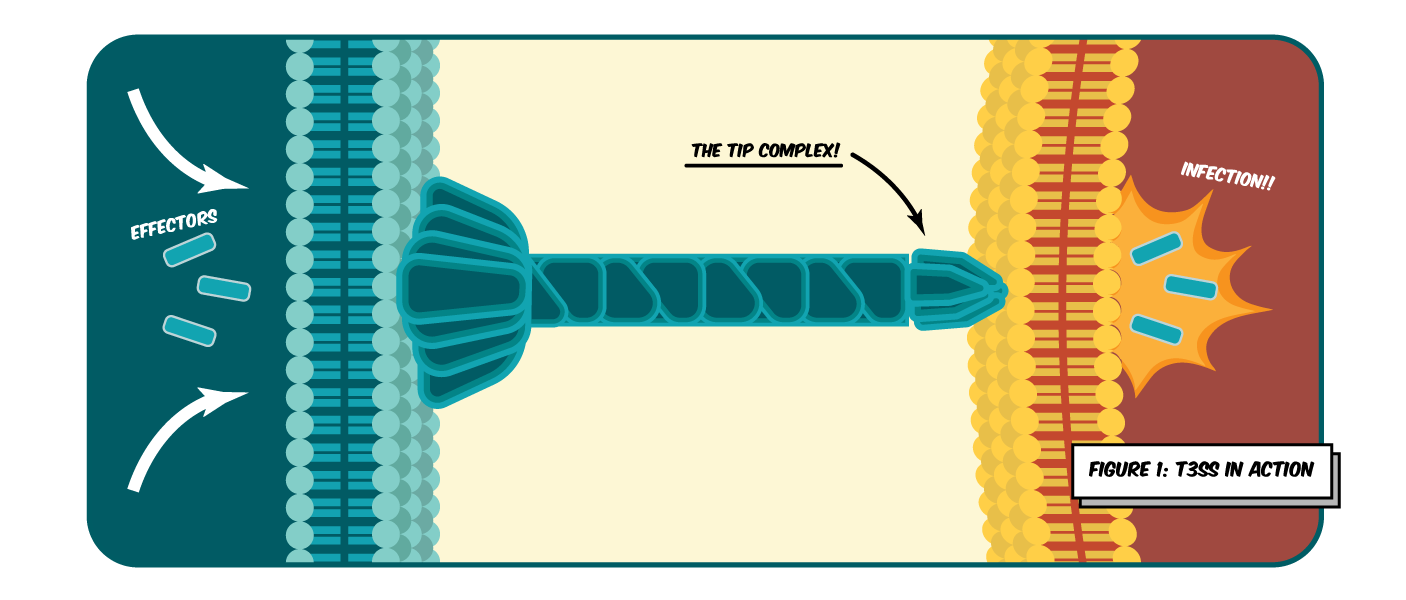
One example, is the Type III Secretion System (T3SS), bacteria’s highly conserved molecular syringe, which self-assembles from over 20 different protein subunits [2]. The T3SS secretes effector proteins directly into host cells. These have a range of activities including cell cycle arrest, membrane remodelling and immune suppression, all which help the bacteria to cause infections and evade the host’s immune system (Figure 1) [3-5].
This results in conditions such as diarrhoea and even more devastating diseases like the black plague and typhoid fever. The tip of the T3SS is a pentameric complex that is required for the insertion of the syringe [6]. Inhibiting the function of the tip proteins has been shown to significantly impair virulence in several pathogenic bacteria [4]. It is also one of the only parts of the T3SS exposed to our immune system [6]. Subsequently, the T3SS tip complex is a primary target for immunisation [7].
Here at Team Injectimod, we're artificially building the T3SS tip complex, which is a new ‘bottom-up’ way of looking at these complex biological machines. To do this, we need the world’s tiniest scaffold! We made this using DNA origami - a method of folding DNA into almost any nano-scale 3D structure. By attaching the protein subunits of the tip to this scaffold, we were able to observe the process of T3SS tip self-assembly for the first time! This will shed light on one of Nature’s greatest mysteries: how self-assembly actually works. It will also allow us to create a novel platform for vaccine development by artificially assembling antigenic structures.