To effectively mimic the assembly of nature’s biomachines, such as the Type 3 secretion system (T3SS) tip complex, requires some structural information about the system.
Therefore, our first task is to review and expand on the current knowledge of the individual proteins the tip complex is assembled from, as well as their stoichiometry and arrangement in complex.
We’ve distilled this information into a set of specifications we call the ‘Biological Blueprint’. This biological blueprint is the basis for the DNA designs that will scaffold the assembly of the T3SS needle tip complex.
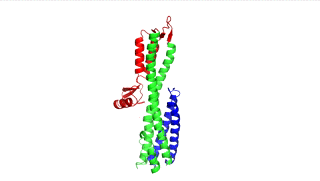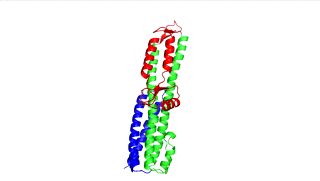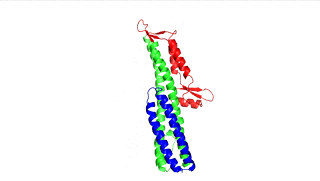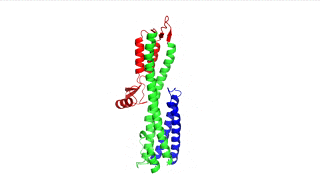
Figure 1: IpaD (PDB 2j0o)

Figure 2: SipD (PDB 2ym9 )
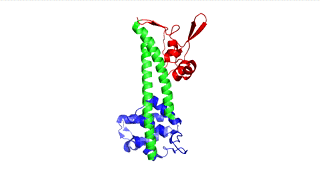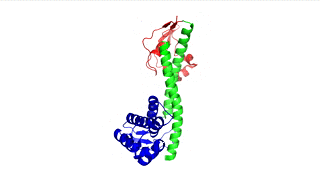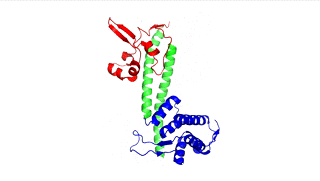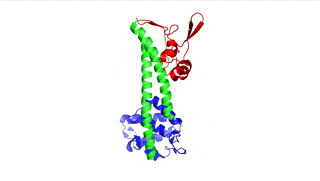
Figure 3: LcrV (PDB 4jbu)
The points of attachment between protein and DNA scaffold are at the termini of the protein sequence, so it is essential that these are accessible. In all three cases, both the N- and C- termini (Figure 4, 5, and 6: purple) are located proximal to the native assembly scaffold the needle filament, and therefore ideal for localizing to a synthetic scaffold.
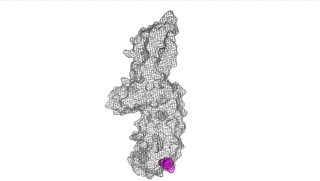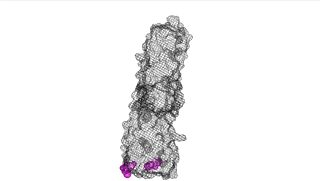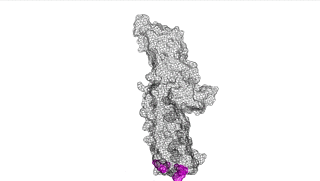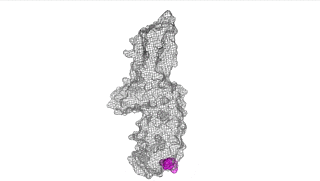
Figure 4: IpaD (PDB 2j0o)
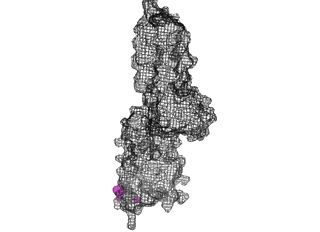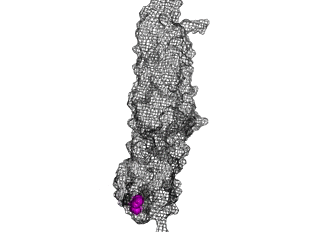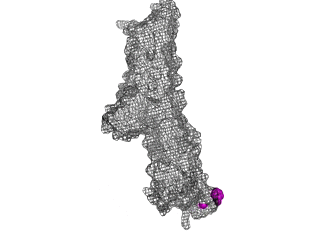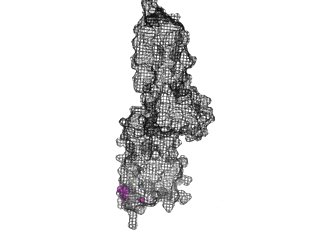
Figure 5: SipD (PDB 2ym9 )
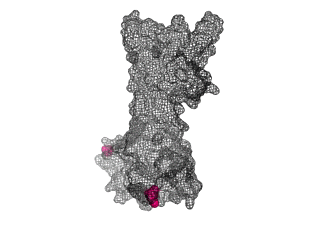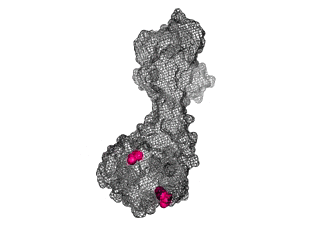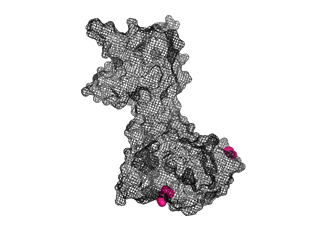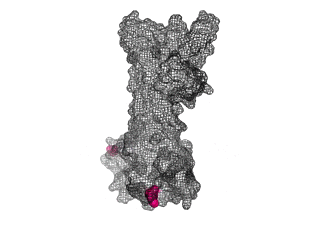
Figure 6: (PDB 4jbu)
The Needle tip complex is generally thought to have pentameric radial symmetry as estimated by STEM [6], crosslinking [7], molecular modeling [1,8], and cryo electron microscopy [7]. It has been suggested that this may be a hetero-pentamer consisting of four needle tip proteins and one ‘translocon’ protein [1, 7]. However bacteria missing the gene for the ‘odd’ translocon protein still form Tip complexes [7], indicating a 5-fold symmetry is possible at the least. We used a pymol script to generate pentameric rings of our protein structures with various orintentations and diameters (Figure 7).
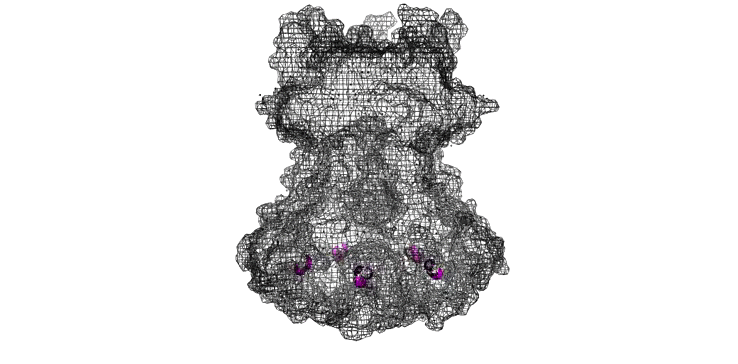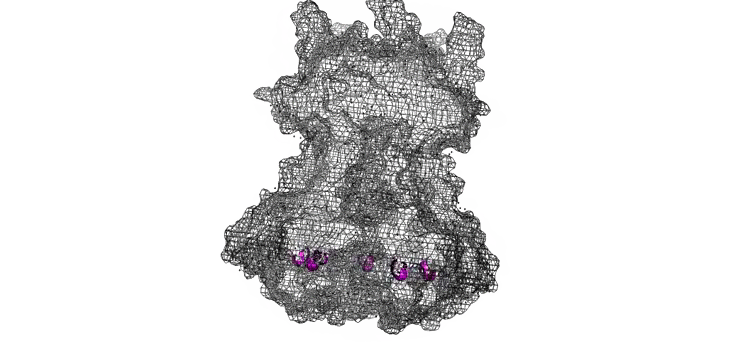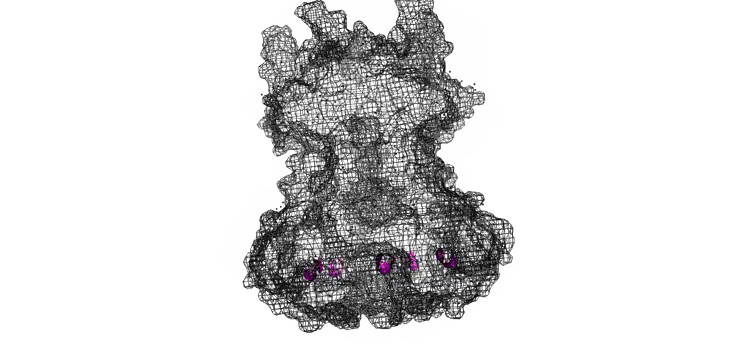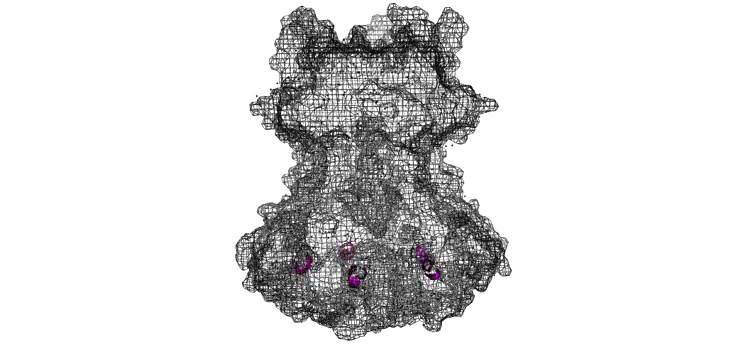
Figure 7: Model of LcrV rotated to have pentameric symmetry
As we are designing an artificial scaffold for assembly, we looked at available models of the natural assembly scaffold for Shigella [4] and Salmonella [5] (Figures 8 and 9 respectively.

Figure 8: Model of Shigella Needle Filament (PDB:2mme)
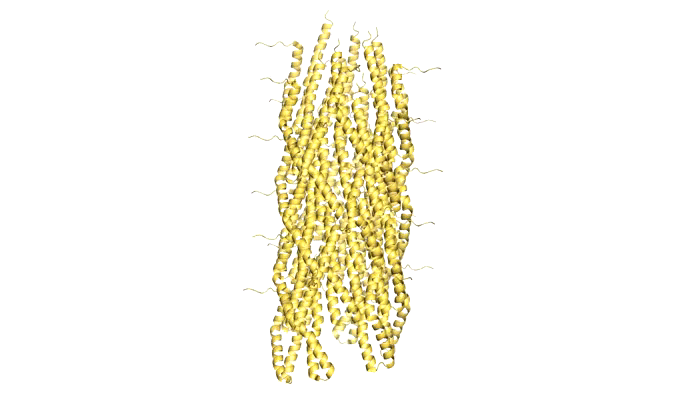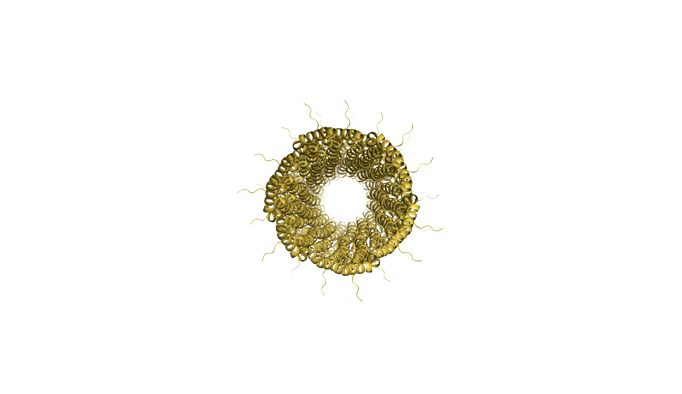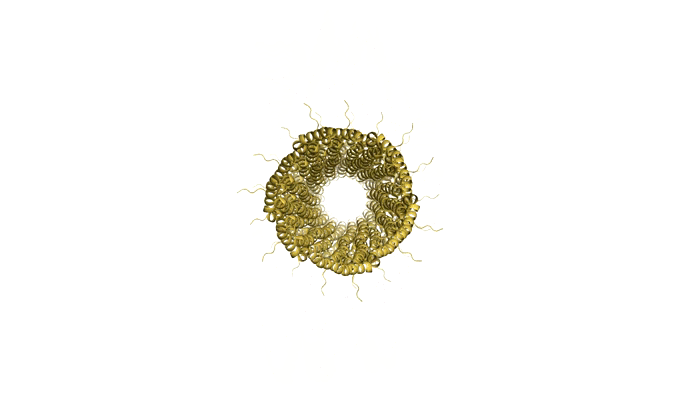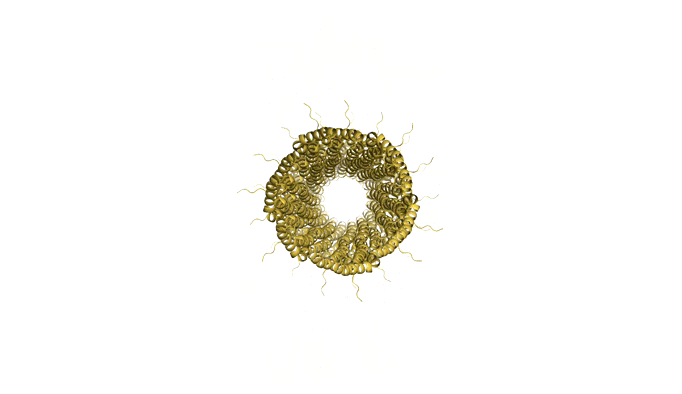
Figure 9: Model of Shigella Needle Filament (PDB:2lpz)
We used the distance function in pymol to measure a diameter range of 60-70Å. This is in agreement with scale of STEM [9] images and in CryoEM reconstructions of the Tip complex on the Filament [7,10].
Finally we generated our own molecular model (Figure 10) of the SipD needle tip complex by docking the needle tip subunits into a calculated electron density map of the tip complex only. We used chimera to subtract the EM density of the needle filiament (EMD:5352) from that of the needle filament and tip complex together (EMD:2801). SipD crystal monomers (3nzz) were then first manually docked into this tip density and the fitted using chimera ‘fit in map’ function.
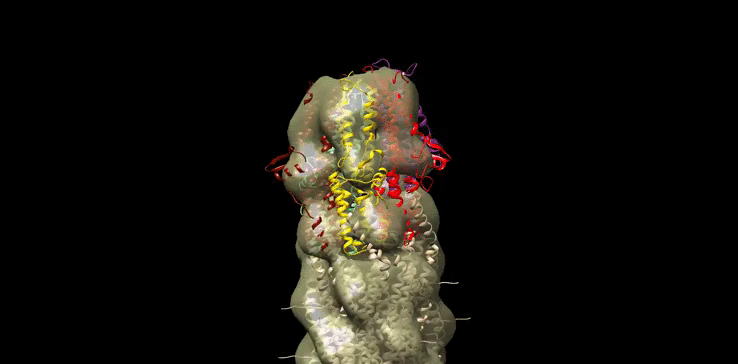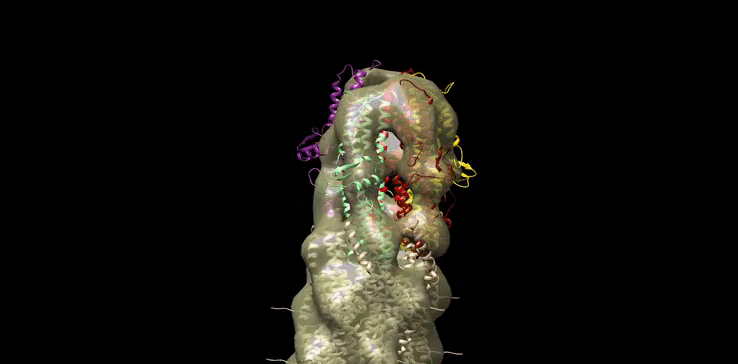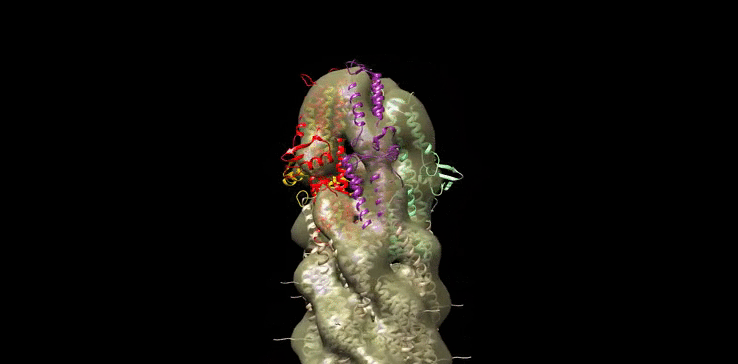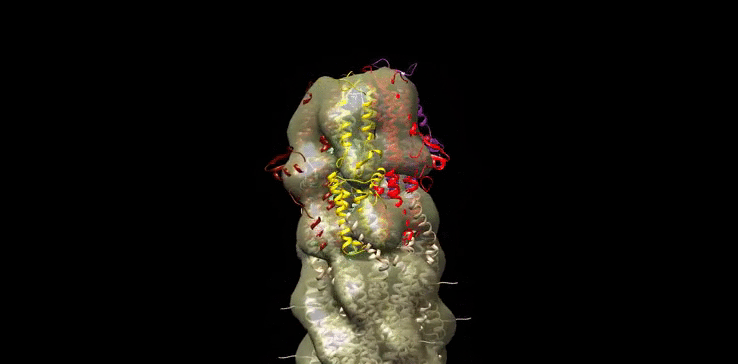
Figure 10: Model of Salmonella Needle tip Complex monomer (PDB:3nzz) docked into Shigella Needle Filament (PDB:2lpz)
After Review the Literature and forming our own molecular models, we now have Biological blueprint that can inform the design of our DNA Scaffolds!!
Now we're ready to DESIGN!!!
[1] Johnson, S., Roversi, P., Espina, M., Olive, A., Deane, J. E., Birket, S., ... & Lea, S. M. (2007). Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. Journal of Biological Chemistry, 282(6), 4035-4044.
[2] Lunelli, M., Hurwitz, R., Lambers, J., & Kolbe, M. (2011). Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog, 7(8), e1002163-e1002163.
[3]Derewenda, U., Mateja, A., Devedjiev, Y., Routzahn, K.M., Evdokimov, A.G., Derewenda, Z.S., and Waugh, D.S. (2004) The structure ofYersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 12: 301–306.
[4]: Demers, J. P., Habenstein, B., Loquet, A., Vasa, S. K., Giller, K., Becker, S., ... & Sgourakis, N. G. (2014). High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nature communications, 5.
[5] Loquet, A., Sgourakis, N. G., Gupta, R., Giller, K., Riedel, D., Goosmann, C., ... & Lange, A. (2012). Atomic model of the type III secretion system needle.Nature, 486(7402), 276-279.
[6] Broz, P., Mueller, C.A., Muller, S.A., Philippsen, A., Sorg, I., Engel, A., and Cornelis, G.R. (2007) Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol 65: 1311–1320.
[7] Cheung, M., Shen, D. K., Makino, F., Kato, T., Roehrich, A. D., Martinez‐Argudo, I., ... & Blocker, A. J. (2015). Three‐dimensional electron microscopy reconstruction and cysteine‐mediated crosslinking provide a model of the type III secretion system needle tip complex. Molecular microbiology, 95(1), 31-50.
[8] Deane, J.E., Roversi, P., Cordes, F.S., Johnson, S., Kenjale, R., Daniell, S., et al. (2006) Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc Natl Acad Sci USA 103: 12529–12533.
[9] Mueller, C. A., Broz, P., Müller, S. A., Ringler, P., Erne-Brand, F., Sorg, I., ... & Cornelis, G. R. (2005). The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science, 310(5748), 674-676.
[10] Epler, C. R., Dickenson, N. E., Bullitt, E., & Picking, W. L. (2012). Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. Journal of molecular biology, 420(1), 29-39.